|
|
|
Welcome to Genes and Development GN434! Contribute something by using the edit button above!
From Genes and Development GN434Evolutionary Differences Between Humans and ChimpsOne of the main differences between Humans and Chimpanzees is the size of the brain, which is larger in human by almost three times, and the development of complex language and higher logical functions. The development of language in humans lead to the spread of idea's and the development of culture and civilizations but the genetic basis of language is still a bit of a mystery. Scientists have isolated a gene known as FOXP2 that they have found to correlate with speech abilities in humans. In individuals with FOXP2 changed through mutations, there is a high risk of developing speech-language disorder 1 (SPCH1) also known as autosomal dominant speech and language disorder with orofacial dyspraxia. This disorder has symptoms of the individual being unable to physically form the correct words using their muscles of their face. FOXP2 is tied to the complex movement and control of the larynx and mouth which is absent in chimpanzees. Point mutations in this gene are tied to severe articulation problems in individuals. It has been determined that individuals need 2 complete and normal FOXP2 for normal language development.
BackgroundFOXP2 is a protein coding gene which is required for the proper development of speech and language. Inactivation and mutation to one allele of this gene causes sever impairments of speech and language which is also known as speech-language disorder (SPCH1), an autosomal dominant speech and language disorder. FOXP2 encodes a member of the forkhead/winged-helix (FOX) family of transcription factors. This transcription factor is 715 amino acids that are highly conserved among mammals, but contains two substitutions at positions 303 and 325. These substitutions occurred after humans diverged from the common ancestor they share with chimpanzees. These two amino acid substitutions are caused by two nucleotide substitutions located on in exon 7 of the FOXP2 gene. Both of these substitutions in exon 7 were found in Neanderthals which made it evident that they were not the cause of the selective sweep of the FOXP2 gene. In order to discover the cause of the selective sweep of the FOXP2 gene experiments were performed in order to find other substitutions. We have been able to identify substitutions in the FOXP2 gene shared by almost all present day humans, but absent it 49,000-year-old Neanderthals by isolating and sequencing genomic DNA fragments. A DNA capture approach was used to isolate the FOXP2 gene from a Neanderthal. The nucleotide sequence of the Neanderthal was compared to multiple present day humans. A substitution found in intron 8 was discovered to affect a certain binding site for the POU3F2 transcription factor. This transcription factor is usually conserved among vertebrates, but it was discovered that the new derived allele in humans is actually less efficient in activating transcription. Since this substitution seems to alter the regulation of the FOXP2 gene and its expression then it could be the cause of the selective sweep.
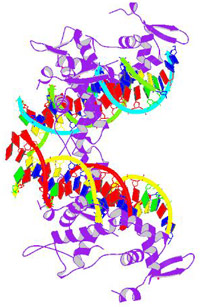
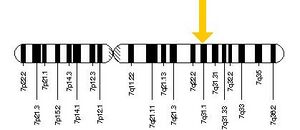
Function of FOXP2The FoxP2 gene plays a role in the development of language and vocalization. Its function was discovered by Vargha-Khadem and a group of her colleagues when they found that the inactivation of FoxP2 led an impairment in developing speech. They discovered its role by observing the behavioral phenotype of the KE family, a family whose half of its members had verbal dyspraxia, a condition in which there is a difficulty in making and co-ordinating movements needed to produce speech. After conducting several tests, the SPCH1 gene was localized, which in turn led to the identification of FOXP2. They found it was located on the long arm of chromosome 7; specifically 7q31. The FoxP2 protein complex contains a FOX DNA binding domain, as well as a large polyglutamine tract. The protein is a transcription factor and interacts with approximately 300 to 400 gene promotors that are located downstream and controls them by repressing their rate of transcription. It is also thought to play a role in the growth of nerve cells and the connections that they make with other cells during the development. FoxP2's location has been described to be in the human embryo between the ages of 6 to 22 weeks. The FoxP2 gene is expressed in various locations of the brain, ranging from the sensory nuclei, limbic nuclei, cerebral cortex and several motor structures, as well as the lungs and the gut. Mutations in the FoxP2 gene have been shown to cause developmental verbal dyspraxia, as shown in the KE family. However, those affected by a mutated FoxP2 show little to no cognitive retardation; they just do not have the motor skills required for speech.
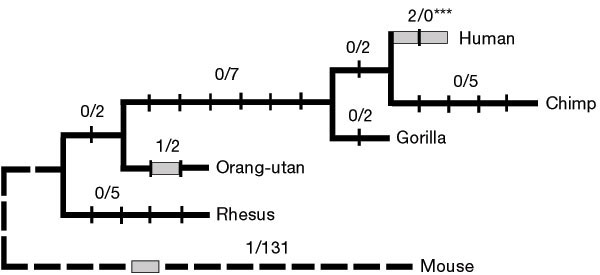
Evolution of FOXP2The FOXP2 gene is a highly conserved, which means that it has very similar DNA sequences in different species and has not evolved much over time. The form of the protein that is encoded by the gene is 715 amino acids long and is the same in the chimpanzee, gorilla and the rhesus monkey. In mice, FoxP2 differs with just one amino acids from these three species. However, in humans the FoxP2 gene differs from the chimpanzee, gorilla and rhesus monkey in two amino acids and three amino acids in comparison with mice. The subtle amino acid changes most likely caused the evolution of language in humans. FOXP2 was concluded to be involved with the brain's ability to learn sequences of movement. Humans specifically use these complex muscle movements to produce sounds for speech. Because Neanderthals had the same FOXP2 gene as modern humans, its implied that they had the capacity for speech and communication. However, evidence shows that they did not have the ability for effective communication and their brains were more adapted towards maximizing their sensory functions.
Experimental ResultsNeanderthal Capture and Sequencing An oligonucleotide array was designed that contained 60-bp-long probes tiled with 3 bp along the FOXP2 gene. It started from 36,942 nucleotides upstream from the transcription start site and ended 127,832 nucleotides downstream from the inferred polyadenylation site. This array was used to capture DNA fragments from a genomic library made from a 49,000 year old Neandertal bone fragment found in Austurias, Spain. Efforts to prevent decontamination with foreign DNA were taken during excavation of the bone and DNA extraction and library construction. Also, the DNA adaptors used to construct the library were tagged so that DNA contamination from outside sources could be detected. The library was then hybridized to the array, washed, and the captured DNA fragments were extracted through washing with a solvent. The fragments were then amplified. A second array was hybridized using the same probes and DNA fragments were extracted through the same method. Again, the fragments were amplified. The DNA fragments were then sequenced from both ends using an Illumina GAII instrument. The readings that came from the same fragment were merged, and fragments with identical end segments were fused. The sequences were then mapped to the human genome. Through this method, about 90% of the positions were covered by at least one Neandertal sequence. To get a more accurate reading, published data from the genomes of three Croatian Neandertals were added, so that about 94% of the FOXP2 region was covered by the genome of an archaic human. 1,669 substitutions that likely occurred in and around the FOXP2 gene in the human evolutionary lineage were extracted by finding the areas that were different from the corresponding position in the chimpanzee, orangutan, and rhesus macaque. 1,415 of these had the derived human-like allele while 143 of them carried the ancestral ape-like allele. At 42 positions it was seen that there was a copy of the derived and the ancestral allele in the archaic DNA. Of these 42, 20 were excluded that only had a substitution of an A or a T. The other 22 had the more derived allele carrying a C or G. These substitutions are probably a result of cytosine residue deamination. Thus, a total of 165 genes remained for analysis. To identify which substitutions were fixed or at high rates in humans, the array from above was used again to isolate and sequence the FOXP2 gene in 50 humans, each from a different population. If a position had available data from 20 or more humans, the derived allele had to be at a frequency of 95% or higher. For the remaining positions, allele counts were added by looking them up in the single nucleotide polymorphism database (dbSNP). Again the frequency had to be 95% or higher. One position did not have any data, indicating it is not variable in humans. 46 derived positions were found to be at a frequency of 95% or higher in humans and were analyzed further. Next, the researchers identified which positions overlapped with functional or conserved regions by looking in the UCSC genome browser. Two positions were found to be significantly conserved among vertebrates. These, along with five other positions, were located within DNAse hypersensitive sites and/or binding sites for transcription factors. One of these positions, position 114076877, was located in a region where a putative sweep occurred, in which variation surrounding a mutation is eliminated by natural selection. This derived base is found on the putatively selected haplotype, while the ancestral base is on the nonselected haplotype. This indicates that this position may be responsible for the selective sweep. Position 114076877 is the 3rd base in the 14-bp long transcription factor binding site for POU3F2. The 1.4-kb region around this position is highly conserved among tetrapods. It carries an adenine residue in all tetrapods and all 3 DNA fragments from the Neandertal from Spain, as well as in the two chromosomes from the Denisovan and in a Neandertal from Siberia. However, in 98% of humans, this adenine residue is replaced with a thymine residue, indicating this position was conserved for 700 million years and only recently changed in humans. The next part of the study focused on whether and how POU3F2 binds to both allelic positions in the putative binding site. To do this, two pairs of oligonucleotides that carry the ancestral and the derived nucleotide were synthesized. HeLa cells were transfected with a POU3F2 expression plasmid and their nuclear extracts were isolated. They were then incubated with one of the two synthesized radioactively labeled oligonucleotides and underwent acryl-amide gel electrophoresis under nonreducing conditions. It was discovered that POU3F2 binds as both a monomer and a dimer at the binding site. However, after performing seven experiments in which the amount of POU3F2 varied it was found that the ratio of dimer to monomer binding was less in the derived allele than in the ancestral allele. Thus, the derived allele binds more of the monomeric form relative to the dimeric form than the ancestral allele. Three pairs of reporter plasmids were then generated to test the extent to which the two variants of POU3F2 binding sites drive transcription. Each plasmid contained either the derived or ancestral allele of the POU3F2 binding site in various configurations with a beta-globin reporter gene. The first plasmid contained two copies of the POU3F2 variant upstream of the reporter gene. The second contained one copy of the POU3F2 variant upstream of four copies of reporter gene. The third contained one copy of the POU3F2 variant upstream of the reporter gene and an SV40 enhancer downstream of the reporter gene. HeLa cells were transfected with separate plasmids with a reference plasmid and a POU3F2 expression if indicated. Two days later, RNA was extracted from the cells, and reporter and reference genes were assayed using an S1 nuclease protection assay. The first two plasmids failed to show any applicable transcriptional activity. In the third plasmid that contained the POU3F2 variant upstream of the SV40 enhancer, however, both the derived and the ancestral allele caused expression of the reporter gene. This expression was codependent on POU3F2 expression, so it’s probably mediated by POU3F2 binding to the binding sites. In all experiments with the three plasmids, cells transfected with the derived allele accumulated less reporter mRNA than cells transfected with the ancestral allele. Thus, the derived allele is less efficient in driving transcription of the reporter gene than the ancestral allele. Another test was then performed to test if the 1.8-kb conserved region around the POU3F2 binding site can drive transcription during mouse development. Transgenic mice were created carrying one of the two variants of the conserved region in front of a minimal promoter and a lacZ reporter gene. Animals and embryos were analyzed for reporter gene expression in the brain, heart, esophagus, and lungs at embryonic day E11.5, and postpartal days P4, P28, and P60. At no time was any reproducible staining detected. Thus, it can be concluded that the conserved region does not act as an enhancer in the mouse at the development stages tested. References1. Ayub, Q. et al. "FOXP2 Targets Show Evidence of Positive Selection in European Populations". The American Journal of Human Genetics. 92:5 (2013): 696-706. Web. 2. Enard, W., Przeworski, M., Fisher, S., Lai, C., Wiebe, V., & Kitano, T. et al. (2002). Molecular evolution of FOXP2, a gene involved in speech and language. Nature, 418(6900), 869-872. http://dx.doi.org/10.1038/nature01025 3. FOXP2 forkhead box P2 [Homo sapiens (human)] - Gene - NCBI. (2016). Ncbi.nlm.nih.gov. Retrieved 15 March 2016, from http://www.ncbi.nlm.nih.gov/gene/93986 4. FOXP2 Location. (2016). Retrieved from http://ghr.nlm.nih.gov/dynamicImages/chromomap/FOXP2.jpeg 5. MacAndrew, Alec. FOXP2 Mutations. 2016. Web. 26 Feb. 2016. 6. MacAndrew, A. (2003). FOXP2 and the Evolution of Language. Evolutionpages.com. Retrieved 10 March 2016, from http://www.evolutionpages.com/FOXP2_language.htm 7. Maricic, T. et al. "A Recent Evolutionary Change Affects A Regulatory Element In The Human FOXP2 Gene". Molecular Biology and Evolution 30.4 (2012): 844-852. Web. 8. National Center for Biotechnology Information. FOXP2 forkhead box P2 [ Homo sapiens (human) ]. Web. 27 Feb. 2016. 9. Vargha-Khadem, F., Gadian, D., Copp, A., & Mishkin, M. (2005). FOXP2 and the neuroanatomy of speech and language. Nature Reviews Neuroscience, 6(2), 131-138. http://dx.doi.org/10.1038/nrn1605 |