|
|
|
Welcome to Genes and Development GN434! Contribute something by using the edit button above!
From Genes and Development GN434IntroductionIn this breakthrough paper published by Nature in 2014, Stelzer et al., a group of scientists at the Hebrew University of Jerusalem illustrated some important aspects of the molecular ongoings underlying Prader-Willi syndrome. The disease, while being the most well studied one associated to parental imprinting defects, was still elusive in terms of its characterization and potential for cure. The findings from Stelzer et al. provide new material to aid in future studies of Prader-Willi syndrome, along with studies of genomic interactions and their evolution.
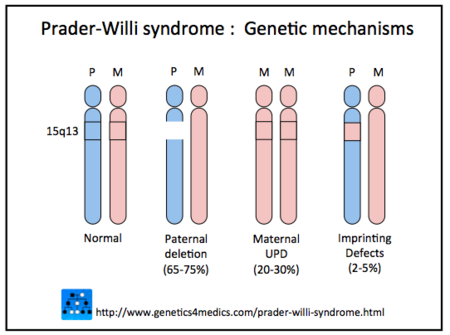
BackgroundPrader-Willi syndrome (PWS) is a genetic disease that is thought to affect the hypothalamus, which helps to control several bodily functions, and so this leads to a wide range of symptoms. The symptoms include varying levels of intellectual disabilities, underdeveloped sex organs or infertiltiy, and metabolic and feeding problems that start in infancy (inability to suck) and persist into adulthood (overeating).1 In 65-70% of cases, Prader-Willi syndrome is caused by a deletion in the PWS region on the paternal copies of a gene on chromosome 152. This gene is normally expressed in a manner called genomic imprinting. Genomic imprinting affects a relatively small amount of genes and causes them to be expressed according to whether they were received from the mother or the father. So, depending on the parent of origin, the copy from the mother is expressed or the copy from the father is expressed, but not both, and the gene that is not expressed is not “activated”or turned off. Paternally expressed genes (PEGs), as are seen in the Prader-Willi gene sequence on chromosome 15, are those that only expressed the genes received from the father, while the genes from the mother are silenced. So when the paternal copies are compromised, no “activated” copy remains to be expressed. When a deletion of the parental copies on chromosome 15 occurs, because the maternal genes are still silenced they cannot make up for the loss, resulting in an overall knockout of the gene function, and by effect Prader- Willi syndrome3. In 20-30% of cases, Prader-Willi syndrome is caused by maternal uniparental disomy of chromosome 152. Uniparental disomy is when someone receives two copies of the gene from the mother3. However, since this gene is one that only expresses paternal copies, both genes are silenced and we get Prader-Willi syndrome. In 1-3% of cases, Prader-Willi is caused by imprinting disorders2. Genes not only can be expressed to show a physical phenotype, but can also be expressed to regulate and repress other genes. As it was found in this study2, genes in the PWS region on chromosome 15 normally function to repress some maternally expressed genes (MEGs) on chromosome 14 using long non-coding RNAs (lncRNAs). Long non-coding RNAs regulate gene expression at the transcriptional and post-transcriptional level. They are molecules that are longer than 2 kb in length and have a coding potential of less than 100 amino acids, and were initially considered non-functional. We have since learned that not only do lncRNA help to regulate normal gene expression but also play a role in the monoallelic expression in genomic imprinting. They also play a role in many other gene functions such as chromatin remodeling, post-transcriptional processing, and intracellular trafficking. Studies are still being done and new information is still being discovered about lncRNAs, because they have such a wide range of functions in a cell4.
FindingsThe work of Stelzer et al. revealed five different layers of information which correlate to a series of studies they conducted1: 1. iPSCs can be successfully modified to model Prader-Willi syndrome. They created four lines of PWS-iPSCs and compared their gene expression to normal/wild-type PSCs, finding that the PEGs in the PWS region of the PWS-iPSCs were completely downregulated (Figure 1d, pictured to the right; the names on the x-axis represent different PEGs in the PWS region). 2. PWS-iPSCs are comparable to parthenogenetic cells (Pg-iPSCs); together, both cell types can be used to isolate gene targets of PEGs in the PWS region. While PWS-iPSCs exhibit downregulation of PEGs due to their being modified and/or silenced, Pg-iPSCs show downregulation because the paternal imprints are completely removed (so the genome is completely maternal) (Figure 1d). Pg-iPSCs can thus not only help uncover imprinted genes, but also those genes’ regulatory targets—the same phenomenon was applied to PWS-iPSCs. Stelzer et al. aimed to identify targets of the PEGs in the PWS region and look for both upregulated and downregulated genes. They found 11 potential target genes that were downregulated, and 12 potential targets that were upregulated. 3. PWS-iPSCs demonstrate altered gene expression in the DLK1-DIO3 locus on chromosome 14. Of the 12 upregulated targets identified, 6 were known MEGs at the imprinted DLK1-DIO3 locus of chromosome 14. This led the researchers to wonder if all the MEGs at the locus would be upregulated; using gene expression analysis, they discovered that all MEG transcripts from the DLK1-DIO3 locus were overexpressed in PWS-iPSCs (Figure 2c, picture to the left; shows upregulation of MEGs in PWS-iPSCs and Pg-iPSCs relative to wild type cells). This solidified the notion that these MEGs are regulatory targets of PEGs from the PWS region. Also, because the PWS-iPSCs were established from unrelated individuals with different causes of disease (some had deletions, some had mUPD), the upregulation of the DLK1-DIO3 MEGs was proven to be a characteristic of Prader-Willi syndrome rather than a random side-occurrence. Stelzer et al. used this discovery to open new doors of inquiry, such as whether the MEGs’ upregulation is dependent on cell type, to which they found that upregulation varies in tissue types and is most noticeable in undifferentiated cells. Another question they pursued was whether the MEGs’ upregulation is directly tied to the symptom of mental retardation observed in PWS patients. Under normal conditions, these MEGs are regularly expressed in the brain’s frontal cortex; their data showed that in PWS brain samples, the MEGs’ expression was even higher. It is important to note that the researchers confirmed their observed results were definitely a byproduct of the MEGs on the maternal chromosome being upregulated, rather than the imprinted paternal chromosome’s DLK1-DIO3 locus being improperly expressed.
4. Expression in the DLK1-DIO3 locus is regulated by IPW. Previous studies have shown that a cluster of the PEGs in the PWS region of chromosome 15 code for long non-coding RNAs (lncRNAs). One of the resulting lncRNAs was identified as IPW (Imprinted in Prader-Willi Syndrome)7. IPW and other lncRNAs regulate gene expression by directing the activity of chromatin modifiers, so Stelzer et al. decided to see if the downregulation of these RNAs in PWS-iPSCs are related to the upregulation of chromosome 14’s MEGs. They overexpressed the lncRNAs from the PWS region PEGs in the PWS cells, and observed that only the expression of IPW significantly downregulated the MEGs at DLK1-DIO3 (Figure 3d, pictured to the right; shows impact of IPW overexpression in Pg and PWS iPSCs).
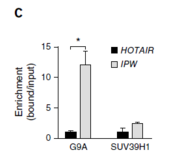
After discovering the impact of IPW’s presence on the expression of the DLK1-DIO3 locus, the researchers wanted to see if this was due to changes in chromatin conformation (as is characteristic of lncRNA regulation). They identified characteristic repressive marks (histone modifications) in the DLK1-DIO3 region with pre-established chromatin maps. Then they used ChIP and qPCR at these mark sites in both normal PSCs and PWS-iPSCs. Comparisons showed that PWS-iPSCs had much lower levels of repressive marks than the normal PSCs. Reintroducing IPW into the PWS cells resulted in these marks being returned to high levels, indicating IPW’s direct involvement. They also tested interactions between IPW and some common histone methyltransferases; IPW was shown to interact in particular with an HMT called G9A, providing further basis for IPW’s role as a regulator of DLK1-DIO3 via chromatin modification (Figure 4c, pictured to left).
Methods/TechniquesInduced Pluripotent Stem Cells Induced pluripotent stem cells (iPSCs) are a specific type of pluripotent stem cell. They differ from other pluripotent stem cells in that they can be generated directly from adult cells and thus do not require unfertilized embryos (parthogenesis embryonic stem cells), fertilized embryos, or embryonic stems cells made by somatic cell nuclear transfer5. iPSCs are made from “reprogrammed” mature cells through the use of viral vectors that introduce specific genes that encode transcription factors. These genes are Oct4, Sox2, cMyc, Klf4, Nanog, and Lin28. The genes encode transcription factors that allow for the specific expression required for the mature cells to become similar to pluripotent embryonic cells.6 iPSCs avoid the many legal and ethical problems associated with ESC isolation from embryos6. iPSCs are useful for personalized healthcare and genetic therapy. This is because the autologous pluripotent cells are obtained from an individual patient's mature cells and avoid issues with hisocompatibiltiy. iPSCs are significantly easier to obtain as the required cells can be obtained at any point in an individual's life.6 This study by Stelzer and his colleagues utilized fibroblasts obtained from two individuals with Prader Willi Syndrome (PWS) and fibroblasts from a third individual with PWS that possessed complete mUPD of chromosome 15. The fibroblast cells were infected with viral particles encoding Oct4, SOX2, KLF4, and MYC. The cells were then grown in embryonic stem cell (ESC) growth conditions and developed into Prader-Willi Syndrome iPSCs (PWS-iPSCs) cultures (examples pictured in Figure 1b). They then used the case-derived PWS-iPSCs from fibroblast cell lines that contained mutations in the corresponding affected region of chromosome 15. In order to compare the PWS-iPSCs and parthenogenetic PSCs (Pg-PSCs) to normal PSCs, they used transcriptome and genome-wide expression analysis.1
Global Gene Expression Analysis In order to study the upregulation of the 12 target genes discovered in the Genome-Wide expression analysis and discern whether the upregulation involved all known MEGs in the DLK1-DIO3 region, a Global micro RNA (miRNA) expression analysis was also conducted. This led to the observation that nearly all MEGs and miRNAs in the specified locus were upregulated more than 3-fold compared to the regulation in normal PSCs. Cells were then obtained from a third individual with Prader-Willis Syndrome who possessed maternal uniparental disomy (mUPD). Another PWS-iPSC line (PWS-iPSC-3) was then generated using these cells. The results of this PWS-iPSC line was then compared to the first two PWS case results to verify that the upregulation of MEGs in the DLK1-DIO3 region was associated with Prader-Willis Syndrome. (Stelzer, Yonatan, Ido Sagi, Ofra Yanuka)
Narrowing Down the True Cause of MEG Upregulation DNA methylation levels at the IG-DMR in both individuals with PWS were studied to support or refute the possible explanation for the upregulation of MEGs in PWS-iPSCs being due to the cell's imprinting abnormalities in the DLK1-DIO3 region independent of their mutations in the PWS region (Figure 2f). The IG-DMR refers to the paternal germline-derived imprinted DMR (differentially methylated regions), and is responsible for this locus being a maternally expressed, monoallelic one.
Studying the Effects of IPW on Chromatin Modifications The researchers performed a chromatin immunoprecipitation analysis followed by quantitative PCR (ChIP-qPCR) at three enrichment sites that were marked by the repressive modifications of histone 3 lysine 27 trimethylation (H3K27me3) and lysine 9 trimethylation (H3K9me3) which were identified in the DLK1 promoter, the IG-DMR, and the region upstream of the snoRNA-113 cluster. They observed enrichment of repressive chromatin marks at all three enrichment sites. The ChIP-qPCR data was then compared across the different cell types and it was observed that significantly lower levels of H3K9me3 marks existed at the IG-DMR in PWS-iPSCs and Pg-iPSCs relative to normal PSCs. The potential protein interactions of IPW were studied to observe how IPW may regulate the expression of MEGs in the DLK1-DIO3 region. (Stelzer, Yonatan, Ido Sagi, Ofra Yanuka)
ImportanceUntil now, very little was known about the underlying causes behind PWS symptoms. Stelzer and his colleagues’ work in this paper has revealed an incredible amount of information about imprinted genes, their normal functionalities, and the detailed impacts of their malfunctioning. Their findings shed light on the mechanisms behind imprinted gene disorders like PWS, and on how imprinted PEGs and MEGs interact and affect one another. In the case of PWS, it is the PEGs on one chromosome repressing the MEGs on another that is so vital to normal organism functioning. The overexpression of the DLK1-DIO3 MEGs in both early tissues and mature brain cells is a new piece of the puzzle that not only provides insight about PWS, but also about the functions of those genes in the DLK1-DIO3 locus on chromosome 14. Most importantly, the fact that the PWS and DLK1-DIO3 loci “cross-talk” lends potential support to theories that deem parental imprinting as an important factor in selecting against asexual reproduction. If paternally and maternally expressed genes are regulated by each other and the organism’s proper functioning depends on those interactions, then sexual reproduction becomes highly selected for. More evidence would be needed to confirm those theories, but for now, Stelzer and his team have shown how broad and complex parental imprinting truly is.
References1. "What Are the Symptoms of Prader-Willi Syndrome (PWS)?" What Are the Symptoms of Prader-Willi Syndrome (PWS)? National Institute of Health, 14 Jan. 2014. Web. 26 Feb. 2016. https://www.nichd.nih.gov/health/topics/prader-willi/conditioninfo/Pages/symptoms.aspx#feeding 2. Stelzer, Yonatan, Ido Sagi, Ofra Yanuka, Rachel Eiges, and Nissim Benvensity. "The Noncoding RNA IPW Regulates the Imprinted DLK1-DIO3 Locus in an Induced Pluripotent Stem Cell Model of Prader-Willi Syndrome." Nature Genetics. Nature, 11 May 2014. Web. 26 Feb. 2016. http://www.nature.com/ng/journal/v46/n6/full/ng.2968.html 3. "What Are Genomic Imprinting and Uniparental Disomy?" Genetics Home Reference. N.p., 22 Feb. 2016. Web. 26 Feb. 2016. https://ghr.nlm.nih.gov/handbook/inheritance/updimprinting 4. Cao, Jinneng. “The Functional Role of Long Non-Coding RNAs and Epigenetics.” Biological Procedures Online 16 (2014): 11. PMC. Web. 27 Feb. 2016. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4177375/ 5. Takahashi, K; Yamanaka, S (2006). "Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors". Cell 126 (4): 663–76 6. A. I. Shevchenko, S. P. Medvedev, N. A. Mazurok, S. M. Zakian (2009). “Induced pluripotent stem cells”. Russian Journal of Genetics 45 (2): 139-146 7. "IPW Gene (RNA Gene) Imprinted In Prader-Willi Syndrome (Non-Protein Coding)." GeneCards: Human Gene Database. Weizmann Institute of Science. Web. 27 Feb. 2016. http://www.genecards.org/cgi-bin/carddisp.pl?gene=IPW |