|
|
|
Welcome to Genes and Development GN434! Contribute something by using the edit button above!
From Genes and Development GN434
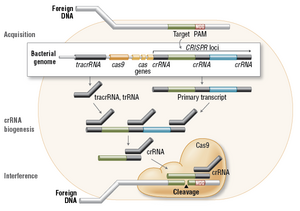
Mouse Muscular Dystrophy and Gene EditingA summary of the original research article: Long, Chengzu, Leonela Amoasii, Alex A. Mireault, John R. McAnally, Hui Li, Efrain Sanchez-Ortiz, Samadrita Bhattacharyya, John M. Shelton, Rhonda Rhonda Bassel-Duby, and Eric N. Olson. "Postnatal Genome Editing Partially Restores Dystrophin Expression in a Mouse Model of Muscular Dystrophy." Science 351.6271 (2016): 400-03. Science. American Association for the Advancement of Science, 22 Jan. 2016. Web. 17 Feb. 2016. OverviewGenome editing in the postnatal stage has proved a potential source to lessen the effects of Duchenne Muscular Dystrophy. In a study completed in mice, scientists found that CRISPR/Cas-9 mediated genome editing could in the future help in a clinical role to treat genetic disease such as DMD. In the study adeno-associate virus-9 (AAV9) was used to get the necessary gene-editing parts to postnatal mdx (organism with DMD). The methods of delivery were altered based on time and modes based on location. Each method proved to restore the dystrophin protein expression in areas affected by disorder such as cardiac and skeletal muscles. The injection increased grip strength of the mice in the weeks following the administration. This study provides hope that gene editing can occur postnatally to correct or lessen the effects of a mutation.  This image shows how DMD affects the muscles of a mouse. A wire test revealed that treating male mdx5Cv mice with tamoxifen for more than a year increased the whole body strength 2- to 3-fold, close to that of normal mice. After exploring the wire for a short period of time (upper right), the mice tried to pull themselves on the wire (bottom left), a very energy-demanding exercise. They would try to pull themselves up until they lost grip with their back feet and hung onto the wire with their forelimbs only (bottom right). From this position, the untreated mdx5Cv mice were unable to sustain their own body weight for more than a few seconds before falling, whereas the mdx5Cv mice treated with tamoxifen (not shown) performed much better and as well as normal mice. Disease ExplainedDuchenne muscular dystrophy (DMD) is a genetic disorder characterized by progressive muscle degeneration and weakness. DMD is a fatal disease caused by mutations in the dystrophin gene, leading to a deficiency of dystrophin protein. Symptoms can begin as early as age 3 in humans. This disease affects 1 in 3500 to 5000 boys. firsts affects the muscles in the hips and pelvic area, as well as the muscles in the thighs and shoulders. Through the development of the disease different muscles begin to become affected. Beginning in the early teens the disease begins to affect the heart and respiratory muscles. This disease was first discovered and described by a French neurologist, Guillaume Benjamin Amand Duchenne. Nothing was known about the genetics cause of DMD until 1987, when the protein associated with this gene was identified and named dystrophin. DMD has an X-linked recessive inheritance pattern and is passed on by the mother. The dystrophin protein contains several domains, including an actin-binding domain at the N terminus, a central rod domain with a series of spectrin-like and actin-binding repeats, and WW and cysteine-rich domains at the C ter- minus that mediate binding to dystroglycan, dystrobrevin, and syntrophin. The actin- binding and cysteine-rich domains are essential for function, but many of the other regions are nonessential. This leads to an estimation that as many as 80% of DMD pateints could benefit from the exon-skipping technique that can skip mutations and those nonessential regions, helping to restore dystrophin expression. Genetic BasisCRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR-associated (Cas) genes are essential in adaptive immunity in select bacteria and archaea, enabling the organisms to respond to and eliminate invading genetic material. Cas9 protein is required for gene silencing The process invloves invading DNA from viruses or plasmids that is cut into small pieces or fragments. These fragments are incorporated into a CRIPSPR locus that is in a gene with a series of short repeats. These get transcribed and then processed to generate small RNAs, a crRNA attached to a CRISPR RNA. Cas9 endonuclease complexed with a crRNA and separate tracrRNA cleaves foreign DNA containing a 20-nucleotide crRNA complementary sequence adjacent to the PAM sequence.
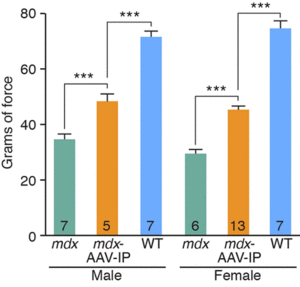
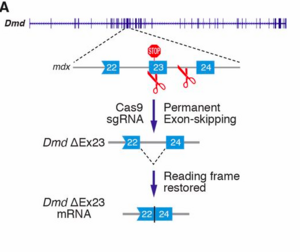
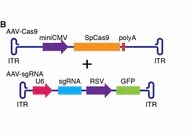
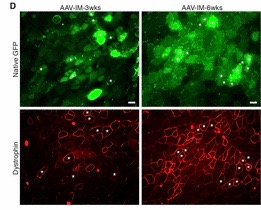
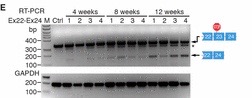
A premature termination codon in exon 23 is responsible for a certain kind of dystrophy in mice. In order to bypass this termination codon, the CRISPR/Cas9 system was used to create NHEJ, which leads to deletions in the gene, which in turn prevents splicing at that exon. Deletions in exons that contain mutations are enough to restore protein expression. The use of NHEJ to fix mutations on the Dmd gene is called myoediting. In order for myoediting to be effective in dealing with the mutation in exon 23, the 5’ and 3’ ends need to be targeted with a specific sgRNA that is directed towards the mutant sequence. Cas9 mRNA and sgRNA-mdx were injected into zygotes without an HDR template, along with either sgRNA-R3 – targets the 3’ end – or sgRNA-L8 – targets the 5’ end. In the resulting progeny, about 80% of mice did not possess exon 23 – this was confirmed through PCR testing. The removal of exon 23 allows for the open reading frame to be revived, this in turn allows dystrophin to be expressed. The restoration of muscle function was observed through grip-strength tests and the measurement of serum creatine kinase levels, which measure muscle membrane permeability. Off-targets effects were also tested for and only 1 of 10 site showed cleavage bands, suggesting that this method of editing is much more efficient than HDR. Scientific Article Images InfoFigure 1B: When applying myoediting to the postnatal tissue, AAV9 was used to display tropism to cardiac and skeletal muscle, which helps deliver Cas9 and sgRNAs to muscles of mice. AAV-guide RNAs were generated by cloning sgRNA-mdx and sgRNA-R3 into AAV-sgRNA vector containing a human U6 promoter and green fluorescent protein(GFP), to have a visual of the inserted AAV9. Figure 1D:Native GFP identified AAV-mediated gene expression in myofibers. Skeletal muscle from the IM-AAV–injected mice showed a mosaic pattern of dystrophin-positive fibers. The percentage of dystrophin-positive myofibers was calculated as a fraction of total estimated fibers, so in the mdx mouse 7.7 ± 3.1% of myofibers tibilis anterior (TA) muscle expressed dystrophin 3 weeks after the injection. Rescue increased to an estimated 25.5 ± 2.9% of myofibers by 6 weeks after injection. Figure 1E: RT- PCR of RNA from Myoedited mdx mice indicates deletion of exon 23. Figure 1F: Sequence of the RT- PCR products of DEx23 band confirmed that exon 22 spliced directly to exon 24, excluding exon 23.
Figure 2: shows the muscle tissues that were analyzed by immunohistochemistry. Image A shows Dystrophin immunostaining of TA muscle is shown for wild-type (WT), mdx, and AAV-RO–treated mdx mice at 4, 8, and 12 weeks after injection. TA muscle of unedited mdx control mice exhibits myonecrosis, indicated by cytoplasm-filling autofluorescence (highlighted with white asterisks). Image B shows dystrophin immunostaining of the heart is illustrated for WT, mdx, and AAV-RO–treated mdx mice at 4, 8, and 12 weeks after injection
References[1] Long, Chengzu, Leonela Amoasii, Alex A. Mireault, John R. McAnally, Hui Li, Efrain Sanchez-Ortiz, Samadrita Bhattacharyya, John M. Shelton, Rhonda Rhonda Bassel-Duby, and Eric N. Olson. "Postnatal Genome Editing Partially Restores Dystrophin Expression in a Mouse Model of Muscular Dystrophy." Science 351.6271 (2016): 400-03. Science. American Association for the Advancement of Science, 22 Jan. 2016. Web. 17 Feb. 2016. [2] Reis, Alex, PhD, Breton Hornblower, PhD, Brett Robb, PhD, and George Tzertzinis, PhD. "CRISPR/Cas9 and Targeted Genome Editing: A New Era in Molecular Biology." NEB Expressions 1 (2014): n. pag. CRISPR/Cas9 and Targeted Genome Editing: A New Era in Molecular Biology. New England Bio Labs Inc., 2014. Web. 13 Mar. 2016. [3] Zubrzycka-Gaarn, E. E., D. E. Bulman, G. Karpati, A. H. Burghes, B. Belfall, H. J. Klamut, J. Talbot, R. S. Hodges, P. N. Ray, and R. G. Worton. "The Duchenne Muscular Dystrophy Gene Product Is Localized in Sarcolemma of Human Skeletal Muscle." Nature 333.6172 (1988): 466-69. Europe PMC. Europe PMC Funders' Groups and Partners. Web. 21 Feb. 2016. [4] "Diseases - DMD - Research." Muscular Dystrophy Association. The Muscular Dystrophy Association, 18 Dec. 2015. Web. 13 Feb. 2016. Return to main page: http://gn434.shoutwiki.com/wiki/Main_Page |